Postdoctoral Research: Integrated food safety systems to reduce foodborne salmonellosis
USDA ARS/SCINet: Meat Safety and Quality Research Unit
Salmonella causes over 1 million illnesses and nearly 400 deaths each year. Despite millions of dollars spent annually and a decrease in Salmonella on meat and poultry products, Salmonella illness rates have not decreased in the last two decades. Our team aims to reduce Salmonella in the meat supply chain by:
1) developing detection assays that are rapid, quantitative, determine serotype, and evaluate virulence;
2) combining data from (1) with whole genome sequencing, metagenomic data, wet lab research, and field studies to generate an understanding of Salmonella population genetics, microbial ecology, and transmission through commodities and their environments;
3) developing effective and affordable pre- and post-harvest interventions for use by commercial producers;
4) integrating data from these studies, data from monitoring programs, and machine learning to create decision support tools for each commodity, production stage, and company;
5) establishing communications and outreach efforts that allow stakeholders to provide feedback to further hone research priorities and facilitate implementation of the program.
I support the team by providing overall project management, data analytics, and data management, and serve as a project leader for Objective 4.
1) developing detection assays that are rapid, quantitative, determine serotype, and evaluate virulence;
2) combining data from (1) with whole genome sequencing, metagenomic data, wet lab research, and field studies to generate an understanding of Salmonella population genetics, microbial ecology, and transmission through commodities and their environments;
3) developing effective and affordable pre- and post-harvest interventions for use by commercial producers;
4) integrating data from these studies, data from monitoring programs, and machine learning to create decision support tools for each commodity, production stage, and company;
5) establishing communications and outreach efforts that allow stakeholders to provide feedback to further hone research priorities and facilitate implementation of the program.
I support the team by providing overall project management, data analytics, and data management, and serve as a project leader for Objective 4.
Doctoral Research: Understanding the amphibian-killing pathogen outside of the amphibian
Briggs Lab. UC Santa Barbara
Fungal pathogens are special-case pathogens which require unique methods for detection, modeling, and management. They have unique characteristics which set them apart from other pathogen groups (bacteria, viruses) and make it difficult to not only accurately detect the pathogen, but estimate its risk to focal hosts and predict disease dynamics. While different in many ways from other pathogen classes, fungal pathogens nonetheless can have similarly devastating impacts on forests, crops, and wildlife. Batrachochytrium dendrobatidis (Bd) is a primary example of a fungal pathogen which has caused devastating global declines and for which new methods are required for detection and prediction. In my PhD thesis, I explored three questions: (1) can we use alternative methods to detect Bd in the environment (i.e., not on amphibians); (2) can we predict Bd in amphibian populations using only information about the amphibian’s habitat; and (3) can Drosophila melanogaster be used as a model organism to study invertebrate-Bd dynamics? By answering these three questions, I aim to elucidate how Bd is able to cause variable responses in amphibian populations (i.e., epidemic vs. endemic states), and understand more about how Bd interacts with its environment as a generalist, fungal pathogen rather than an amphibian specialist.
Check out the Briggs Lab.
Check out the Briggs Lab.
Undergraduate Research: Mapping the spatial distribution of Batrachochytrium salamandrivorans
Zellmer Lab. Occidental College, Los Angeles, California
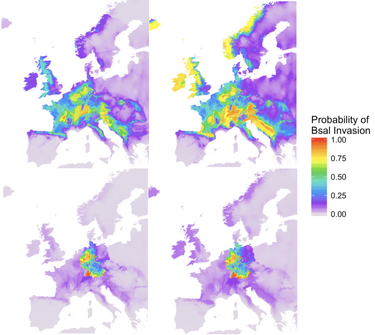 Which of these models of spread best predicts current Bsal presence and absence data?
Which of these models of spread best predicts current Bsal presence and absence data?
Model selection with biased data (i.e., invasive species)
Bsal is thought to be endemic to East Asia, but it is currently causing declines in species naïve to it in Western Europe. Because of this, Bsal is not only an emergent pathogen but also an invasive species. A useful tool to predict the spread of invasive species, called Species Distribution Modeling (SDM), typically uses limited, presence/absence data and machine learning algorithms to identify suitable habitat for a species. Powerful, right? Only if the models can be tested against empirical data to ensure their accuracy! Many tools - model selection techniques - exist to test these models to ensure biological relevance. However, with an invasive species, data can be distributed based on spread dynamics (rather than where habitat is actually suitable), which can ultimately bias the model selection techniques as well. Therefore, is there a model selection technique that handles this biased data better than others? Can we use this technique to identify a model that can predict where Bsal will spread?
This question became the focus of my undergraduate research, and you can read more about it here!
Check out the Zellmer Lab.
Bsal is thought to be endemic to East Asia, but it is currently causing declines in species naïve to it in Western Europe. Because of this, Bsal is not only an emergent pathogen but also an invasive species. A useful tool to predict the spread of invasive species, called Species Distribution Modeling (SDM), typically uses limited, presence/absence data and machine learning algorithms to identify suitable habitat for a species. Powerful, right? Only if the models can be tested against empirical data to ensure their accuracy! Many tools - model selection techniques - exist to test these models to ensure biological relevance. However, with an invasive species, data can be distributed based on spread dynamics (rather than where habitat is actually suitable), which can ultimately bias the model selection techniques as well. Therefore, is there a model selection technique that handles this biased data better than others? Can we use this technique to identify a model that can predict where Bsal will spread?
This question became the focus of my undergraduate research, and you can read more about it here!
Check out the Zellmer Lab.
|
Chytrid survey with the USGS
While Bd is currently widespread across North America, Bsal has yet to make it over here. The USGS is currently swabbing salamanders around North America in the hopes of detecting Bsal as soon as it arrives. My lab and I aided in these efforts and helped to swab Slender Salamanders and California Newts across urban Los Angeles! |
Publications
- Katz, TS; Harhay, DM; Schmidt, JW, Wheeler, TL (2024). Identifying a list of Salmonella serotypes of concern to target for reducing risk of salmonellosis. Frontiers in Microbiology 15:1307563. doi: 10.3389/fmicb.2024.1307563.
- Zellmer, AJ; Slezak, P; Katz, TS. (2020). Clearing up the Crystal Ball: Understanding Uncertainty in Future Climate Suitability Projections for Amphibians. Herpetologica 1 June 2020; 76 (2): 108–120. doi: https://doi.org/10.1655/0018-0831-76.2.108
- Katz, TS & Zellmer AJ.(2018). Comparison of model selection technique performance in predicting the spread of newly invasive species: a case study with Batrachochytrium salamandrivorans. Biological Invasions. https://doi.org/10.1007/s10530-018-1690-7

